Which Of The Following Is Used To Separate Fragments Of Dna By Size?
 Gel electrophoresis apparatus – an agarose gel is placed in this buffer-filled box and an electric electric current is applied via the power supply to the rear. The negative final is at the far end (black wire), so Dna migrates toward the positively charged anode (red wire). | |
| Classification | Electrophoresis |
|---|---|
| Other techniques | |
| Related | Capillary electrophoresis SDS-PAGE 2-dimensional gel electrophoresis Temperature slope gel electrophoresis |

The epitome above shows how modest DNA fragments will migrate through agarose quickly but large size DNA fragments move more slowly during electrophoresis. The graph to the right shows the nonlinear human relationship between the size of the Deoxyribonucleic acid fragment and the distance migrated.

Gel electrophoresis is a process where an electric current is practical to Deoxyribonucleic acid samples creating fragments that can be used for comparison between DNA samples.
one) DNA is extracted.
2) Isolation and distension of Deoxyribonucleic acid.
3) Dna added to the gel wells.
iv) Electrical current applied to the gel.
5) DNA bands are separated by size.
6) Dna bands are stained.
Gel electrophoresis is a method for separation and analysis of macromolecules (Dna, RNA and proteins) and their fragments, based on their size and charge. Information technology is used in clinical chemistry to separate proteins by accuse or size (IEF agarose, essentially size independent) and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins past charge.[1]
Nucleic acid molecules are separated by applying an electric field to move the negatively charged molecules through a matrix of agarose or other substances. Shorter molecules move faster and migrate farther than longer ones considering shorter molecules migrate more easily through the pores of the gel. This phenomenon is called sieving.[2] Proteins are separated by the accuse in agarose considering the pores of the gel are as well small to sieve proteins. Gel electrophoresis can as well exist used for the separation of nanoparticles.
Gel electrophoresis uses a gel as an anticonvective medium or sieving medium during electrophoresis, the motility of a charged particle in an electrical current. Gels suppress the thermal convection acquired by the application of the electric field, and can besides human action as a sieving medium, slowing the passage of molecules; gels can also but serve to maintain the finished separation so that a post electrophoresis stain can be applied.[3] Dna Gel electrophoresis is usually performed for belittling purposes, ofttimes later amplification of DNA via polymerase chain reaction (PCR), but may be used as a preparative technique prior to utilise of other methods such as mass spectrometry, RFLP, PCR, cloning, DNA sequencing, or Southern blotting for farther characterization.
Concrete basis [edit]
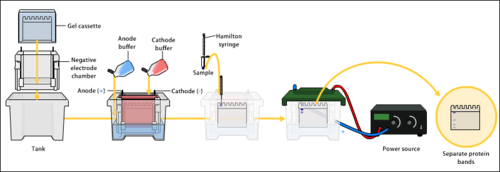
Overview of gel electrophoresis.
Electrophoresis is a procedure that enables the sorting of molecules based on size. Using an electric field, molecules (such as Deoxyribonucleic acid) can exist made to motion through a gel made of agarose or polyacrylamide. The electric field consists of a negative charge at i finish which pushes the molecules through the gel, and a positive charge at the other end that pulls the molecules through the gel. The molecules existence sorted are dispensed into a well in the gel material. The gel is placed in an electrophoresis chamber, which is and so connected to a power source. When the electric field is applied, the larger molecules move more than slowly through the gel while the smaller molecules move faster. The different sized molecules course distinct bands on the gel.[4]
The term "gel" in this instance refers to the matrix used to contain, and so separate the target molecules. In virtually cases, the gel is a crosslinked polymer whose limerick and porosity are chosen based on the specific weight and composition of the target to be analyzed. When separating proteins or small nucleic acids (DNA, RNA, or oligonucleotides) the gel is commonly equanimous of different concentrations of acrylamide and a cantankerous-linker, producing different sized mesh networks of polyacrylamide. When separating larger nucleic acids (greater than a few hundred bases), the preferred matrix is purified agarose. In both cases, the gel forms a solid, nonetheless porous matrix. Acrylamide, in dissimilarity to polyacrylamide, is a neurotoxin and must be handled using appropriate safety precautions to avoid poisoning. Agarose is equanimous of long unbranched chains of uncharged carbohydrates without cantankerous-links resulting in a gel with big pores allowing for the separation of macromolecules and macromolecular complexes.[5]
Electrophoresis refers to the electromotive force (EMF) that is used to motility the molecules through the gel matrix. By placing the molecules in wells in the gel and applying an electrical field, the molecules will move through the matrix at different rates, determined largely by their mass when the accuse-to-mass ratio (Z) of all species is uniform. However, when charges are non all uniform the electrical field generated by the electrophoresis procedure volition cause the molecules to migrate differentially according to charge. Species that are net positively charged volition migrate towards the cathode which is negatively charged (because this is an electrolytic rather than galvanic cell), whereas species that are cyberspace negatively charged will migrate towards the positively charged anode. Mass remains a factor in the speed with which these not-uniformly charged molecules migrate through the matrix toward their respective electrodes.[6]
If several samples have been loaded into adjacent wells in the gel, they will run parallel in private lanes. Depending on the number of different molecules, each lane shows the separation of the components from the original mixture as one or more than distinct bands, one ring per component. Incomplete separation of the components tin can lead to overlapping bands, or indistinguishable smears representing multiple unresolved components.[ citation needed ] Bands in unlike lanes that terminate up at the aforementioned altitude from the top contain molecules that passed through the gel at the same speed, which usually means they are approximately the aforementioned size. There are molecular weight size markers bachelor that incorporate a mixture of molecules of known sizes. If such a marker was run on i lane in the gel parallel to the unknown samples, the bands observed can be compared to those of the unknown to determine their size. The altitude a band travels is approximately inversely proportional to the logarithm of the size of the molecule (alternatively, this can exist stated as the distance traveled is inversely proportional to the log of samples's molecular weight).[7]
There are limits to electrophoretic techniques. Since passing a current through a gel causes heating, gels may cook during electrophoresis. Electrophoresis is performed in buffer solutions to reduce pH changes due to the electric field, which is of import because the charge of DNA and RNA depends on pH, simply running for besides long tin can frazzle the buffering capacity of the solution. There are also limitations in determining the molecular weight by SDS-Folio, especially when trying to notice the MW of an unknown poly peptide. Certain biological variables are difficult or impossible to minimize and can bear upon electrophoretic migration. Such factors include protein structure, post-translational modifications, and amino acid composition. For example, tropomyosin is an acidic protein that migrates abnormally on SDS-PAGE gels. This is because the acidic residues are repelled by the negatively charged SDS, leading to an inaccurate mass-to-charge ratio and migration.[8] Further, unlike preparations of genetic material may non migrate consistently with each other, for morphological or other reasons.
Types of gel [edit]
The types of gel most typically used are agarose and polyacrylamide gels. Each type of gel is well-suited to different types and sizes of the analyte. Polyacrylamide gels are usually used for proteins and have very high resolving power for small-scale fragments of DNA (v-500 bp). Agarose gels, on the other mitt, take lower resolving power for DNA but have a greater range of separation, and are therefore used for DNA fragments of usually 50–20,000 bp in size, but the resolution of over 6 Mb is possible with pulsed field gel electrophoresis (PFGE).[9] Polyacrylamide gels are run in a vertical configuration while agarose gels are typically run horizontally in a submarine mode. They also differ in their casting methodology, as agarose sets thermally, while polyacrylamide forms in a chemic polymerization reaction.
Agarose [edit]

Inserting the gel comb in an agarose gel electrophoresis chamber
Agarose gels are fabricated from the natural polysaccharide polymers extracted from seaweed. Agarose gels are hands cast and handled compared to other matrices because the gel setting is a physical rather than chemical alter. Samples are also easily recovered. After the experiment is finished, the resulting gel can be stored in a plastic handbag in a fridge.
Agarose gels do not have a uniform pore size, but are optimal for electrophoresis of proteins that are larger than 200 kDa.[10] Agarose gel electrophoresis can as well exist used for the separation of Deoxyribonucleic acid fragments ranging from 50 base pair to several megabases (millions of bases)[ citation needed ], the largest of which require specialized apparatus. The distance between DNA bands of different lengths is influenced by the pct agarose in the gel, with higher percentages requiring longer run times, sometimes days. Instead high percent agarose gels should exist run with a pulsed field electrophoresis (PFE), or field inversion electrophoresis.
"About agarose gels are made with between 0.7% (skilful separation or resolution of large v–10kb Dna fragments) and ii% (practiced resolution for small 0.2–1kb fragments) agarose dissolved in electrophoresis buffer. Upwards to 3% can be used for separating very tiny fragments but a vertical polyacrylamide gel is more than appropriate in this case. Low pct gels are very weak and may interruption when you endeavour to lift them. High pct gels are frequently brittle and do not set evenly. 1% gels are mutual for many applications."[11]
Polyacrylamide [edit]
Polyacrylamide gel electrophoresis (PAGE) is used for separating proteins ranging in size from 5 to ii,000 kDa due to the uniform pore size provided by the polyacrylamide gel. Pore size is controlled by modulating the concentrations of acrylamide and bis-acrylamide pulverization used in creating a gel. Care must exist used when creating this type of gel, every bit acrylamide is a potent neurotoxin in its liquid and powdered forms.
Traditional Dna sequencing techniques such as Maxam-Gilbert or Sanger methods used polyacrylamide gels to separate DNA fragments differing past a unmarried base-pair in length so the sequence could exist read. Most mod DNA separation methods now use agarose gels, except for particularly pocket-size DNA fragments. It is currently most often used in the field of immunology and protein analysis, often used to separate different proteins or isoforms of the same protein into split bands. These can be transferred onto a nitrocellulose or PVDF membrane to exist probed with antibodies and corresponding markers, such as in a western blot.
Typically resolving gels are fabricated in half dozen%, 8%, 10%, 12% or 15%. Stacking gel (5%) is poured on superlative of the resolving gel and a gel comb (which forms the wells and defines the lanes where proteins, sample buffer, and ladders volition be placed) is inserted. The percentage called depends on the size of the protein that one wishes to identify or probe in the sample. The smaller the known weight, the higher the percentage that should be used. Changes in the buffer system of the gel can help to farther resolve proteins of very small sizes.[12]
Starch [edit]
Partially hydrolysed potato starch makes for another non-toxic medium for protein electrophoresis. The gels are slightly more than opaque than acrylamide or agarose. Non-denatured proteins tin be separated according to charge and size. They are visualised using Napthal Black or Amido Black staining. Typical starch gel concentrations are five% to 10%.[xiii] [14] [15]
Gel conditions [edit]
Denaturing [edit]

TTGE profiles representing the bifidobacterial diverseness of fecal samples from two good for you volunteers (A and B) before and after AMC (Oral Amoxicillin-Clavulanic Acid) treatment
Denaturing gels are run nether conditions that disrupt the natural structure of the analyte, causing it to unfold into a linear concatenation. Thus, the mobility of each macromolecule depends only on its linear length and its mass-to-charge ratio. Thus, the secondary, 3rd, and quaternary levels of biomolecular structure are disrupted, leaving only the master structure to be analyzed.
Nucleic acids are oft denatured by including urea in the buffer, while proteins are denatured using sodium dodecyl sulfate, usually as function of the SDS-Page process. For full denaturation of proteins, it is also necessary to reduce the covalent disulfide bonds that stabilize their third and fourth structure, a method called reducing Folio. Reducing weather are usually maintained by the addition of beta-mercaptoethanol or dithiothreitol. For a general assay of protein samples, reducing PAGE is the almost common form of poly peptide electrophoresis.
Denaturing conditions are necessary for proper estimation of molecular weight of RNA. RNA is able to form more intramolecular interactions than Deoxyribonucleic acid which may effect in modify of its electrophoretic mobility. Urea, DMSO and glyoxal are the nearly often used denaturing agents to disrupt RNA construction. Originally, highly toxic methylmercury hydroxide was often used in denaturing RNA electrophoresis,[16] merely it may be method of choice for some samples.[17]
Denaturing gel electrophoresis is used in the Deoxyribonucleic acid and RNA banding pattern-based methods temperature gradient gel electrophoresis (TGGE)[18] and denaturing slope gel electrophoresis (DGGE).[19]
Native [edit]

Native gels are run in non-denaturing weather so that the analyte's natural structure is maintained. This allows the physical size of the folded or assembled complex to affect the mobility, allowing for assay of all four levels of the biomolecular structure. For biological samples, detergents are used only to the extent that they are necessary to lyse lipid membranes in the cell. Complexes remain—for the most office—associated and folded every bit they would be in the cell. 1 downside, however, is that complexes may not split up cleanly or predictably, as it is difficult to predict how the molecule'south shape and size will touch its mobility. Addressing and solving this problem is a major aim of quantitative native Page.
Unlike denaturing methods, native gel electrophoresis does not use a charged denaturing amanuensis. The molecules beingness separated (usually proteins or nucleic acids) therefore differ non only in molecular mass and intrinsic charge, simply also the cantankerous-exclusive surface area, and thus experience different electrophoretic forces dependent on the shape of the overall structure. For proteins, since they remain in the native state they may exist visualized not only past general protein staining reagents but also by specific enzyme-linked staining.
A specific experiment example of an application of native gel electrophoresis is to check for enzymatic activity to verify the presence of the enzyme in the sample during protein purification. For example, for the protein alkaline phosphatase, the staining solution is a mixture of 4-chloro-2-2methylbenzenediazonium table salt with 3-phospho-2-naphthoic acid-two'-iv'-dimethyl aniline in Tris buffer. This stain is commercially sold as a kit for staining gels. If the poly peptide is present, the machinery of the reaction takes identify in the following society: it starts with the de-phosphorylation of 3-phospho-2-naphthoic acid-2'-4'-dimethyl aniline by alkaline metal phosphatase (water is needed for the reaction). The phosphate group is released and replaced by an alcohol group from water. The electrophile 4- chloro-2-2 methylbenzenediazonium (Fast Crimson TR Diazonium salt) displaces the booze group forming the final production Red Azo dye. Equally its proper noun implies, this is the final visible-red production of the reaction. In undergraduate academic experimentation of protein purification, the gel is usually run adjacent to commercial purified samples to visualize the results and conclude whether or not purification was successful.[21]
Native gel electrophoresis is typically used in proteomics and metallomics. However, native Folio is also used to scan genes (DNA) for unknown mutations every bit in Single-strand conformation polymorphism.
Buffers [edit]
Buffers in gel electrophoresis are used to provide ions that carry a current and to maintain the pH at a relatively constant value. These buffers accept plenty of ions in them, which is necessary for the passage of electricity through them. Something like distilled water or benzene contains few ions, which is not ideal for the apply in electrophoresis.[22] At that place are a number of buffers used for electrophoresis. The most mutual beingness, for nucleic acids Tris/Acetate/EDTA (TAE), Tris/Borate/EDTA (TBE). Many other buffers have been proposed, e.one thousand. lithium borate, which is rarely used, based on Pubmed citations (LB), isoelectric histidine, pK matched goods buffers, etc.; in virtually cases the purported rationale is lower current (less heat) matched ion mobilities, which leads to longer buffer life. Borate is problematic; Borate can polymerize, or collaborate with cis diols such every bit those found in RNA. TAE has the lowest buffering capacity merely provides the best resolution for larger Dna. This ways a lower voltage and more time, merely a better product. LB is relatively new and is ineffective in resolving fragments larger than five kbp; Nonetheless, with its low conductivity, a much higher voltage could exist used (up to 35 V/cm), which means a shorter analysis time for routine electrophoresis. As low as ane base pair size difference could be resolved in iii% agarose gel with an extremely low conductivity medium (one mM Lithium borate).[23]
Most SDS-Page poly peptide separations are performed using a "discontinuous" (or DISC) buffer system that significantly enhances the sharpness of the bands within the gel. During electrophoresis in a discontinuous gel organisation, an ion gradient is formed in the early stage of electrophoresis that causes all of the proteins to focus on a single precipitous band in a process called isotachophoresis. Separation of the proteins by size is achieved in the lower, "resolving" region of the gel. The resolving gel typically has a much smaller pore size, which leads to a sieving effect that now determines the electrophoretic mobility of the proteins.
Visualization [edit]
After the electrophoresis is complete, the molecules in the gel can exist stained to brand them visible. DNA may exist visualized using ethidium bromide which, when intercalated into DNA, fluoresce under ultraviolet light, while protein may be visualised using silver stain or Coomassie brilliant blue dye. Other methods may also be used to visualize the separation of the mixture's components on the gel. If the molecules to be separated contain radioactivity, for example in a DNA sequencing gel, an autoradiogram can be recorded of the gel. Photographs tin can be taken of gels, often using a Gel Doc system.
Downstream processing [edit]
After separation, an boosted separation method may and so be used, such every bit isoelectric focusing or SDS-PAGE. The gel will then exist physically cut, and the protein complexes extracted from each portion separately. Each extract may then be analysed, such as by peptide mass fingerprinting or de novo peptide sequencing after in-gel digestion. This can provide a great bargain of information about the identities of the proteins in a complex.
Applications [edit]
- Interpretation of the size of Dna molecules following restriction enzyme digestion, e.g. in restriction mapping of cloned Deoxyribonucleic acid.
- Analysis of PCR products, e.1000. in molecular genetic diagnosis or genetic fingerprinting
- Separation of restricted genomic DNA prior to Southern transfer, or of RNA prior to Northern transfer.
Gel electrophoresis is used in forensics, molecular biology, genetics, microbiology and biochemistry. The results can be analyzed quantitatively by visualizing the gel with UV lite and a gel imaging device. The image is recorded with a calculator-operated camera, and the intensity of the ring or spot of interest is measured and compared confronting standard or markers loaded on the aforementioned gel. The measurement and analysis are mostly done with specialized software.
Depending on the type of assay being performed, other techniques are often implemented in conjunction with the results of gel electrophoresis, providing a broad range of field-specific applications.
Nucleic acids [edit]
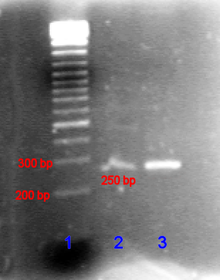
An agarose gel of a PCR production compared to a DNA ladder.
In the instance of nucleic acids, the management of migration, from negative to positive electrodes, is due to the naturally occurring negative charge carried by their carbohydrate-phosphate backbone.[24]
Double-stranded DNA fragments naturally behave as long rods, so their migration through the gel is relative to their size or, for circadian fragments, their radius of gyration. Circular DNA such equally plasmids, nevertheless, may evidence multiple bands, the speed of migration may depend on whether it is relaxed or supercoiled. Unmarried-stranded DNA or RNA tends to fold up into molecules with complex shapes and migrate through the gel in a complicated manner based on their tertiary construction. Therefore, agents that disrupt the hydrogen bonds, such equally sodium hydroxide or formamide, are used to denature the nucleic acids and cause them to comport as long rods again.[25]
Gel electrophoresis of big Dna or RNA is usually done past agarose gel electrophoresis. See the "Chain termination method" page for an example of a polyacrylamide Dna sequencing gel. Label through ligand interaction of nucleic acids or fragments may exist performed by mobility shift affinity electrophoresis.
Electrophoresis of RNA samples tin be used to check for genomic DNA contamination and besides for RNA degradation. RNA from eukaryotic organisms shows distinct bands of 28s and 18s rRNA, the 28s band existence approximately twice as intense every bit the 18s band. Degraded RNA has less sharply defined bands, has a smeared advent, and the intensity ratio is less than 2:one.
Proteins [edit]
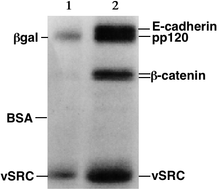
SDS-Folio autoradiography – The indicated proteins are present in different concentrations in the two samples.
Proteins, unlike nucleic acids, tin have varying charges and complex shapes, therefore they may not migrate into the polyacrylamide gel at similar rates, or all when placing a negative to positive EMF on the sample. Proteins, therefore, are usually denatured in the presence of a detergent such as sodium dodecyl sulfate (SDS) that coats the proteins with a negative charge.[iii] Mostly, the amount of SDS leap is relative to the size of the protein (ordinarily 1.4g SDS per gram of protein), so that the resulting denatured proteins have an overall negative charge, and all the proteins have a similar charge-to-mass ratio. Since denatured proteins act like long rods instead of having a complex tertiary shape, the rate at which the resulting SDS coated proteins drift in the gel is relative only to its size and non its charge or shape.[3]
Proteins are normally analyzed past sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-Folio), by native gel electrophoresis, by preparative gel electrophoresis (QPNC-Page), or by ii-D electrophoresis.
Characterization through ligand interaction may be performed past electroblotting or by affinity electrophoresis in agarose or by capillary electrophoresis equally for estimation of bounden constants and determination of structural features like glycan content through lectin binding.
Nanoparticles [edit]
A novel application for gel electrophoresis is to split or characterize metal or metallic oxide nanoparticles (east.g. Au, Ag, ZnO, SiO2) regarding the size, shape, or surface chemistry of the nanoparticles. The telescopic is to obtain a more than homogeneous sample (e.chiliad. narrower particle size distribution), which then can exist used in further products/processes (due east.g. self-assembly processes). For the separation of nanoparticles within a gel, the particle size about the mesh size is the primal parameter, whereby ii migration mechanisms were identified: the unrestricted mechanism, where the particle size << mesh size, and the restricted mechanism, where particle size is similar to mesh size.[26]
History [edit]
- 1930s – first reports of the use of sucrose for gel electrophoresis
- 1955 – introduction of starch gels, mediocre separation (Smithies)[14]
- 1959 – introduction of acrylamide gels; disc electrophoresis (Ornstein and Davis); accurate command of parameters such every bit pore size and stability; and (Raymond and Weintraub)
- 1966 – outset use of agar gels[27]
- 1969 – introduction of denaturing agents especially SDS separation of protein subunit (Weber and Osborn)[28]
- 1970 – Laemmli separated 28 components of T4 phage using a stacking gel and SDS
- 1972 – agarose gels with ethidium bromide stain[29]
- 1975 – 2-dimensional gels (O'Farrell); isoelectric focusing then SDS gel electrophoresis
- 1977 – sequencing gels
- 1983 – pulsed field gel electrophoresis enables separation of big Dna molecules
- 1983 – introduction of capillary electrophoresis
- 2004 – introduction of a standardized time of polymerization of acrylamide gels enables make clean and predictable separation of native proteins (Kastenholz)[xxx]
A 1959 book on electrophoresis past Milan Bier cites references from the 1800s.[31] Notwithstanding, Oliver Smithies made pregnant contributions. Bier states: "The method of Smithies ... is finding broad application considering of its unique separatory power." Taken in context, Bier clearly implies that Smithies' method is an comeback.
See also [edit]
- History of electrophoresis
- Electrophoretic mobility shift assay
- Gel extraction
- Isoelectric focusing
- Pulsed field gel electrophoresis
- Nonlinear frictiophoresis
- Two-dimensional gel electrophoresis
- SDD-AGE
- Zymography
- Fast parallel proteolysis[32]
References [edit]
- ^ Kryndushkin DS, Alexandrov IM, Ter-Avanesyan Physician, Kushnirov VV (2003). "Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104". J Biol Chem. 278 (49): 49636–43. doi:x.1074/jbc.M307996200. PMID 14507919.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sambrook, Joseph (2001). Molecular cloning : a laboratory transmission (in Spanish). Cold Spring Harbor, Due north.Y: Common cold Spring Harbor Laboratory Press. ISBN978-0-87969-576-7. OCLC 45015638.
- ^ a b c Berg, Jeremy (2002). Biochemistry (in Estonian). New York: Due west.H. Freeman. ISBN978-0-7167-4955-4. OCLC 48055706.
- ^ Wilson, Keith (2018). Wilson and Walker'southward principles and techniques of biochemistry and molecular biology. Cambridge, United Kingdom New York, NY: Cambridge University Press. ISBN978-1-316-61476-1. OCLC 998750377.
- ^ Boyer, Rodney (2000). Modern experimental biochemistry (in Estonian). San Francisco: Benjamin Cummings. ISBN978-0-8053-3111-0. OCLC 44493241.
- ^ Robyt, John (1990). Biochemical techniques : theory and practice. Prospect Heights, Ill: Waveland Printing. ISBN978-0-88133-556-9. OCLC 22549624.
- ^ Lee PY, Costumbrado J, Hsu CY, Kim YH (2012). "Agarose gel electrophoresis for the separation of Dna fragments". J Vis Exp (62). doi:ten.3791/3923. PMC4846332. PMID 22546956.
{{cite periodical}}: CS1 maint: multiple names: authors listing (link) - ^ "Molecular Weight Determination past SDS-Folio, Rev B" (PDF). www.bio-rad.com . Retrieved 23 March 2022.
- ^ Tom Maniatis; Due east. F. Fritsch; Joseph Sambrook (1982). "Chapter 5, protocol 1". Molecular Cloning - A Laboratory Manual. Vol. 1 (3rd ed.). p. 5.2–five.3. ISBN978-0879691363.
- ^ Smisek, David L.; Hoagland, David A. (1989). "Agarose gel electrophoresis of high molecular weight, constructed polyelectrolytes". Macromolecules. American Chemical Lodge (ACS). 22 (5): 2270–2277. Bibcode:1989MaMol..22.2270S. doi:x.1021/ma00195a048. ISSN 0024-9297.
- ^ "Agarose gel electrophoresis (bones method)". Biological Protocols . Retrieved 23 March 2022.
- ^ Schägger H (2006). "Tricine-SDS-Page". Nat Protoc. i (ane): 16–22. doi:10.1038/nprot.2006.iv. PMID 17406207. S2CID 209529082.
- ^ Gordon, A.H. (1969). Electrophoresis of Proteins in Polyacrylamide and Starch Gels: Laboratory technique in Biochemistry and Molecular Biology. Amsterdam: Due north-The netherlands Pub. Co. ISBN978-0-7204-4202-1. OCLC 21766.
- ^ a b Smithies O (1955). "Zone electrophoresis in starch gels: group variations in the serum proteins of normal human adults". Biochem J. 61 (4): 629–41. doi:ten.1042/bj0610629. PMC1215845. PMID 13276348.
- ^ Wraxall BG, Culliford BJ (1968). "A thin-layer starch gel method for enzyme typing of bloodstains". J Forensic Sci Soc. eight (2): 81–2. doi:10.1016/s0015-7368(68)70449-seven. PMID 5738223.
- ^ Buell GN, Wickens MP, Payvar F, Schimke RT (1978). "Synthesis of full length cDNAs from four partially purified oviduct mRNAs". J Biol Chem. 253 (7): 2471–82. doi:x.1016/S0021-9258(17)38097-3. PMID 632280.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Schelp C, Kaaden OR (1989). "Enhanced full-length transcription of Sindbis virus RNA by effective denaturation with methylmercury hydroxide". Acta Virol. 33 (3): 297–302. PMID 2570517.
- ^ Fromin North, Hamelin J, Tarnawski S, Roesti D, Jourdain-Miserez Thousand, Forestier N; et al. (2002). "Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns". Environ Microbiol. iv (11): 634–43. doi:ten.1046/j.1462-2920.2002.00358.x. PMID 12460271.
{{cite journal}}: CS1 maint: multiple names: authors listing (link) - ^ Fischer SG, Lerman LS (1979). "Length-independent separation of Dna restriction fragments in 2-dimensional gel electrophoresis". Cell. 16 (ane): 191–200. doi:10.1016/0092-8674(79)90200-nine. PMID 369706. S2CID 9369012.
- ^ Hempelmann Eastward, Wilson RJ (1981). "Detection of glucose-6-phosphate dehydrogenase in malarial parasites". Mol Biochem Parasitol. 2 (3–4): 197–204. doi:x.1016/0166-6851(81)90100-vi. PMID 7012616.
- ^ Ninfa AJ, Ballou DP (1998). Fundamental Approaches to Biochemistry and Biotechnology. Bethesda, Doc: Fitzgerald Science Press. ISBN9781891786006.
- ^ Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (2009). fundamental laboratory approaches for biochemistry and biotechnology. Hoboken, NJ: Wiley. p. 161. ISBN978-0470087664.
- ^ Brody JR, Kern SE (2004). "History and principles of conductive media for standard DNA electrophoresis". Anal Biochem. 333 (1): one–13. doi:10.1016/j.ab.2004.05.054. PMID 15351274.
- ^ Lodish H; Berk A; Matsudaira P (2004). Molecular Jail cell Biology (fifth ed.). WH Freeman: New York, NY. ISBN978-0-7167-4366-8.
- ^ Troubleshooting DNA agarose gel electrophoresis. Focus 19:3 p.66 (1997).
- ^ Barasinski, Matthäus; Garnweitner, Georg (12 February 2020). "Restricted and Unrestricted Migration Mechanisms of Silica Nanoparticles in Agarose Gels and Their Utilization for the Separation of Binary Mixtures". The Journal of Physical Chemistry C. American Chemical Society (ACS). 124 (9): 5157–5166. doi:ten.1021/acs.jpcc.9b10644. ISSN 1932-7447. S2CID 213566317.
- ^ Thorne HV (1966). "Electrophoretic separation of polyoma virus Deoxyribonucleic acid from host cell DNA". Virology. 29 (2): 234–9. doi:10.1016/0042-6822(66)90029-8. PMID 4287545.
- ^ Weber K, Osborn M (1969). "The reliability of molecular weight determinations past dodecyl sulfate-polyacrylamide gel electrophoresis". J Biol Chem. 244 (16): 4406–12. doi:x.1016/S0021-9258(18)94333-iv. PMID 5806584.
- ^ Aaij C, Borst P (1972). "The gel electrophoresis of Dna". Biochim Biophys Acta. 269 (ii): 192–200. doi:10.1016/0005-2787(72)90426-1. PMID 5063906.
- ^ Kastenholz, Bernd (2004). "Preparative Native Continuous Polyacrylamide Gel Electrophoresis (PNC‐Folio): An Efficient Method for Isolating Cadmium Cofactors in Biological Systems". Belittling Messages. Informa UK Express. 37 (iv): 657–665. doi:10.1081/al-120029742. ISSN 0003-2719. S2CID 97636537.
- ^ Bier, Milan (1959). Electrophoresis: theory, methods, and applications. Academic Printing. p. 225. OCLC 1175404.
- ^ Minde, David P.; Maurice, Madelon 1000.; Rüdiger, Stefan M. D. (3 October 2012). Uversky, Vladimir N. (ed.). "Determining Biophysical Poly peptide Stability in Lysates by a Fast Proteolysis Analysis, FASTpp". PLOS ONE. Public Library of Science (PLoS). 7 (x): e46147. Bibcode:2012PLoSO...746147M. doi:10.1371/journal.pone.0046147. ISSN 1932-6203. PMC3463568. PMID 23056252.
External links [edit]
- Biotechniques Laboratory electrophoresis demonstration, from the University of Utah's Genetic Science Learning Center
- Discontinuous native poly peptide gel electrophoresis
- Drinking harbinger electrophoresis
- How to run a Dna or RNA gel
- Animation of gel analysis of DNA brake
- Stride past step photos of running a gel and extracting DNA
- A typical method from wikiversity
Which Of The Following Is Used To Separate Fragments Of Dna By Size?,
Source: https://en.wikipedia.org/wiki/Gel_electrophoresis
Posted by: edgeswitithe77.blogspot.com


0 Response to "Which Of The Following Is Used To Separate Fragments Of Dna By Size?"
Post a Comment